Tell me who your buddies are ....
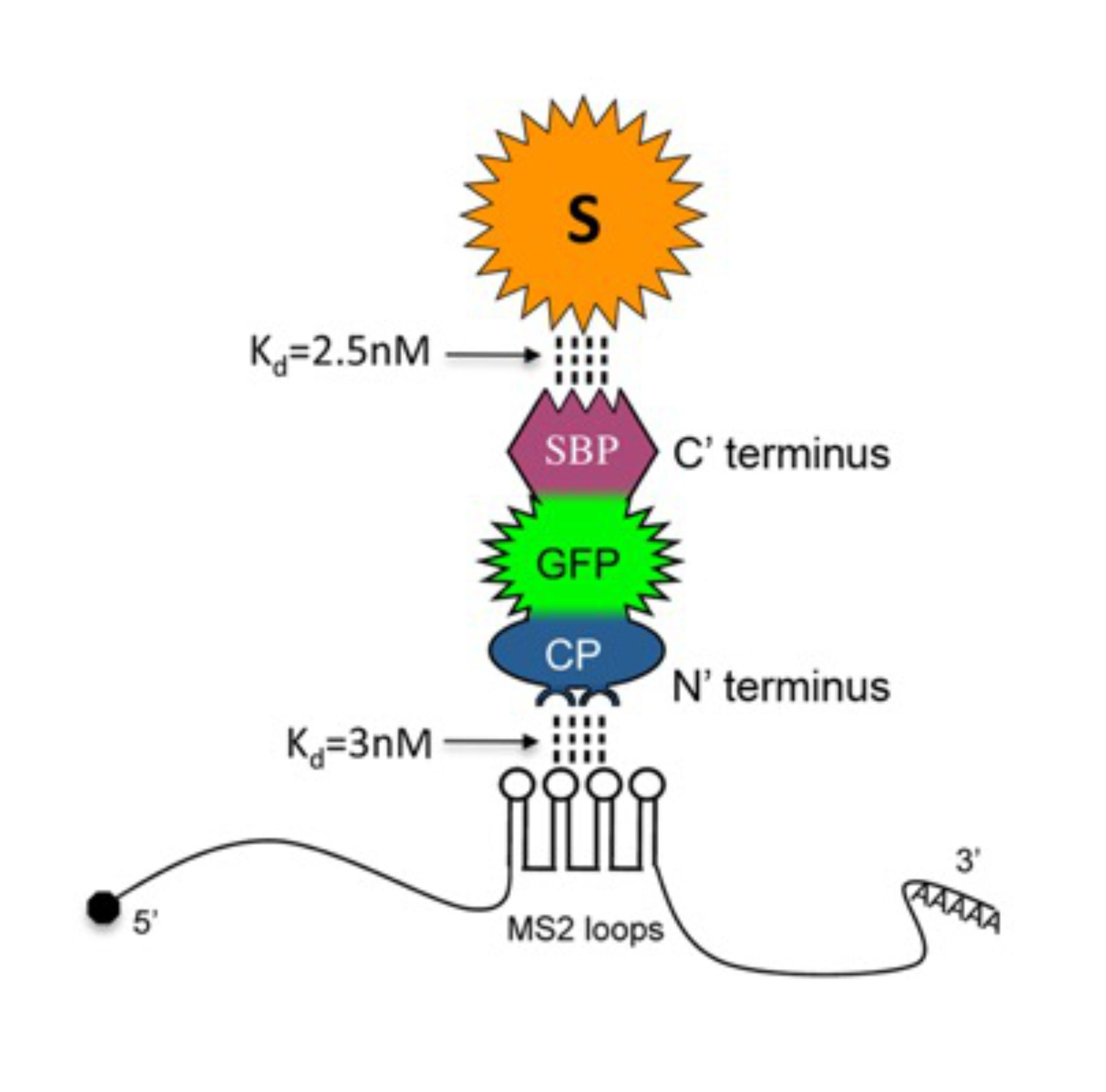
The RNA in the cells is never alone. It is always accompanied by multiple proteins that take care and protect it from degradation. The RNA-protein interactions are typically transient, meaning that any mRNA may interact with dozens of proteins throughout its life. But how can we dissect the intrinsic net of RNA-protein interactions? To address this challenge, we established a novel method for direct isolation of mRNAs of interest from living cells. This was done using a chimeric protein, CP-GFP-SBP. The CP moiety recognizes the specific secondary RNA structure, called MS2 loops, embedded within the RNA of interest. The GFP domain enables visualization of the RNA of interest. The SBP (streptavidin-binding peptide) possesses very high affinity to streptavidin, allowing for biochemical isolation. This simple and diverse method, named RaPID (for RNA-binding protein purification and identification), enabled visualization of RNAs of interest in the living cells, purification of mRNA-protein complexes with the ability to subsequently analyze both proteins and RNAs that interact with the mRNAs of interest. This method further helped to characterize the interaction between mRNAs and translation factors, identify factors responsible for mitochondrial delivery of nuclear-encoded mRNAs, and identify multiplexed assembly of mRNAs in yeast. Here you can find the protocol of the RaPID procedure.
Excuse me, how do I get to the mitochondrion?
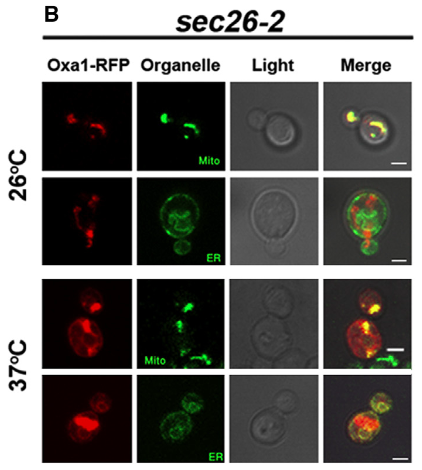
Eukaryotic cells are compartmentalized, meaning that they have distinct spaces within the cells. There are few such compartments named organelles, such as endoplasmic reticulum (ER), Golgi, mitochondrion. Each one of these organelles has a different protein composition. Taken into account that the majority of these proteins are encoded by nuclear genes, how these proteins find their way to the particular organelle? We have addressed this question by studying how mRNAs encoding for mitochondrial proteins are transported to the mitochondrion. Using RaPID methodology, we found that the COPI complex, known to regulate vesicles shuttling between the Golgi and ER, is responsible for the mitochondrial localization of nuclear-encoded mRNAs. When the integrity of COPI was compromised, mitochondrial mRNAs were mislocalized to the ER. As a result, mitochondrial proteins were also mislocalized to ER. Therefore, COPI is important for the proper localization of mitochondrial proteins and, as a result, for adequate mitochondrial functioning.
Brothers in arms
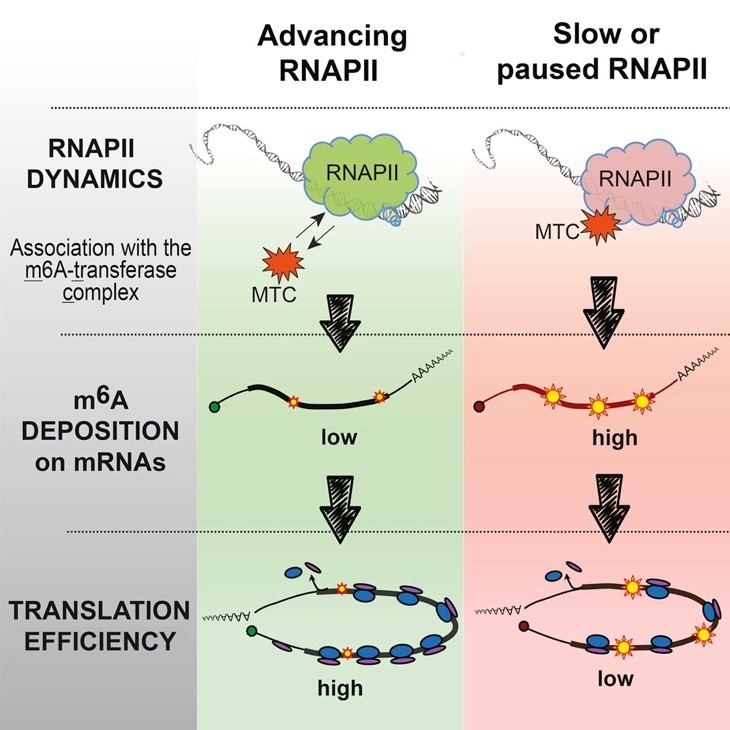
Transcription and translation are two major stages of gene expression in all living cells. Because these processes are integral parts of the gene expression cascade, one could assume that their coordination is important. However, transcription and translation exist at different space and time, making the question of coordination between them intriguing. Our study, performed at the Netherlands Cancer Institute in Amsterdam, found that nuclear transcription of DNA to mRNA can impact the efficiency of mRNA translation to proteins. Simply put, genes efficiently transcribed into mRNAs tend also to be more efficiently translated into proteins. We found that this effect can be mediated by the co-transcriptional addition of methyl group to adenosines in mRNAs, known as m6A modification. Inefficient transcription often leads to enhanced deposition of m6A, which, in turn, reduces the translation dynamics when located within open reading frame. Thus, the m6A mark bridges the nuclear transcription with cytosolic translation.
So close no matter how far...
Following the observation that nuclear transcription can impact cytosolic translation, we decided to further investigate the connection between these processes. We screened a lot of literature and, remarkably, found multiple evidence showing that these two steps of the gene expression cascade indeed have multiple ties connecting them. To establish the unifying model of molecular pathways bridging transcription and translation, we wrote a review paper. The first part of its name was taken from the timeless words of the song “Nothing else matters” by Metallica, symbolizing the intricate connections between the seemingly different fields as art, poetry, music and science.
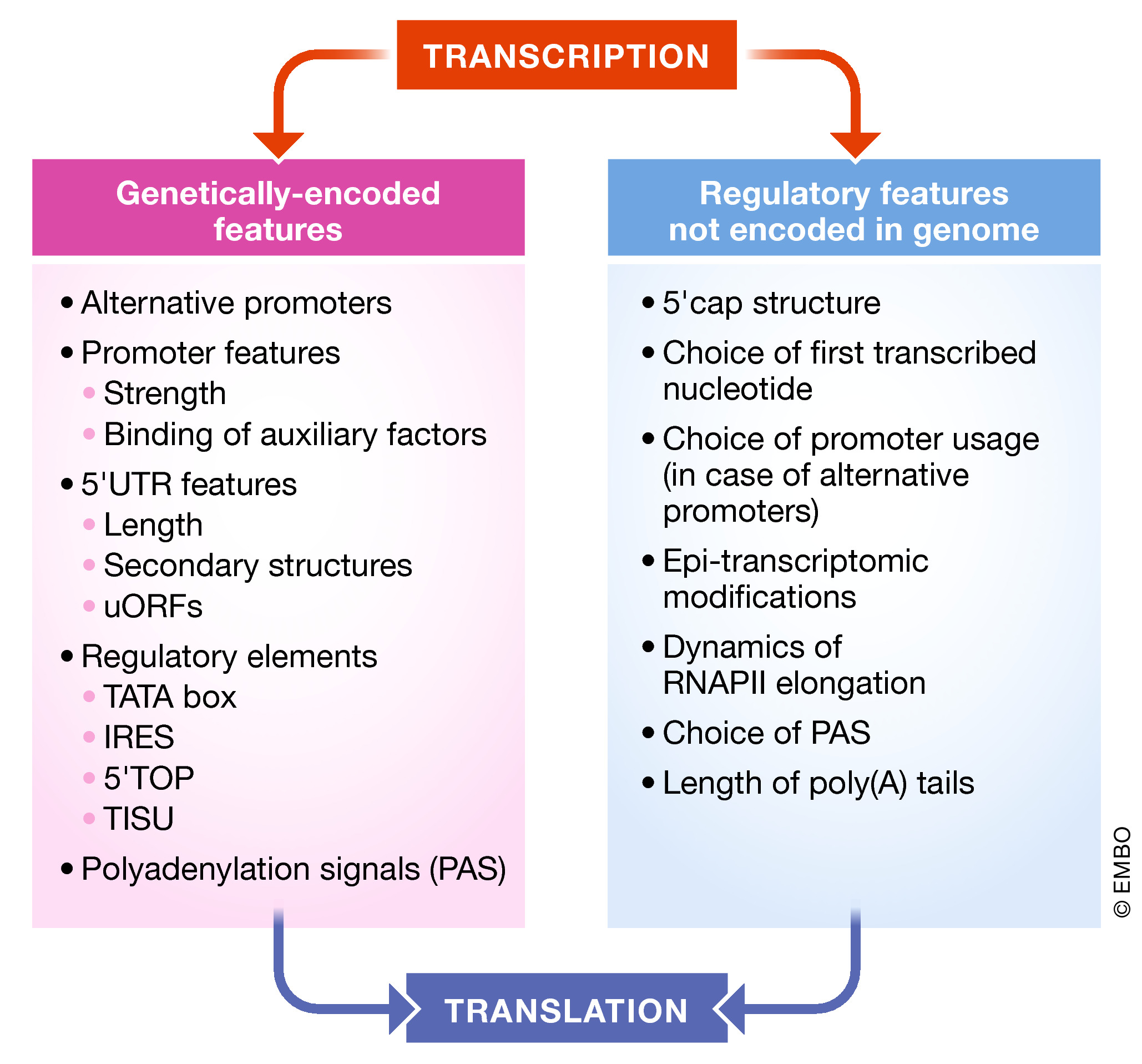
Tell me how you were born and I will tell when you shall die
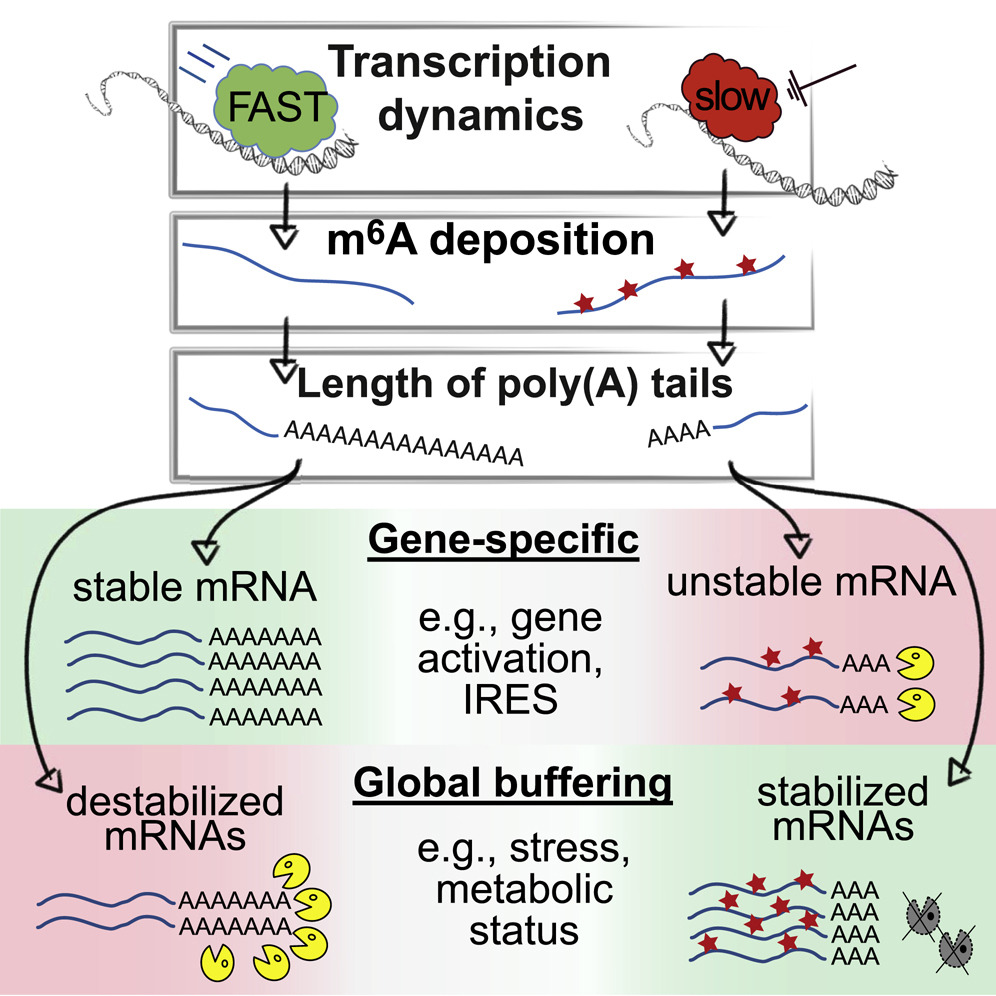
Does it sound weird if during the process of RNA synthesis, the timing of its death would be pre-determined? Given the ability of transcription to regulate translation, we hypothesized that also the timing of RNA degradation can be affected by the transcription process. To test this idea, we employed biochemical, genomic, and epi-transcriptomic approaches and demonstrated that mRNAs transcription of which has been compromised, tend to have shorter half-life. This effect was due to shortened poly(A) tail that, in turn, was affected by enhanced levels of m6A on these transcripts. However, quite contra-intuitively, we found that global impediment of transcription led to a dramatic stabilization of almost all (97%!) endogenous mRNAs. When we sought for a reason, we found that in these conditions, proteins that degrade mRNAs are repressed, which leads to a remarkable stabilization (T1/2 of >24 hours) of mRNAs. Therefore, both findings strongly indicated that cellular transcription can regulate mRNA degradation.
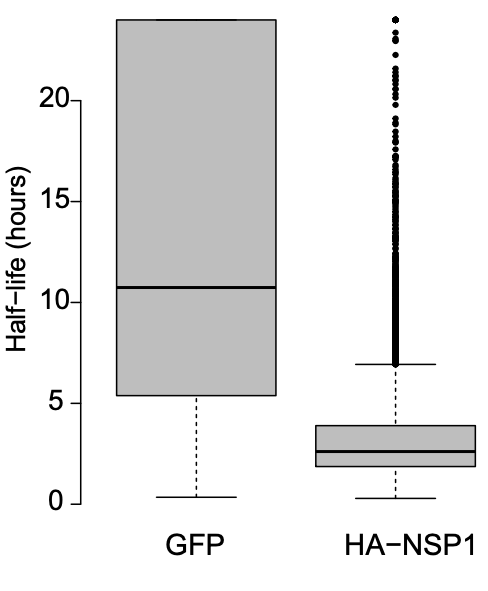
RNA regulation by SARS-CoV-2
Following the outbreak of COVID-19, we decided to help study the RNA regulation of SARS-CoV-2 to fight the pandemic. The genome of SARS-CoV-2 is a huge RNA molecule with a very complex regulation, and we wished to understand out how this virus promotes the expression of its own genes while inhibiting the host gene expression. We found that that the first expressed viral protein, NSP1, induces degradation of most mRNAs of its host, in parallel to boosting the expression of the viral transcripts. Searching for the element that protect viral mRNAs, we have identified a specific RNA element that protects viral transcripts and enhances their expression. This element is somewhat surprising: it simply lacks guanosines near the 5’cap structure, and following this simple rule allows even non-viral genes to be efficiently expressed in the presence of NSP1. Additionally, we found that the genomic version of 5’UTR of SARS-CoV-2 can mediate cap-independent protein translation, a feature that is common to multiple positive-stranded RNA viruses. Cumulatively, this study has unveiled a few “tricks” that help to repress host genes and boost the expression of viral transcripts.